For Science 🔭#
Why I Choose to Use Python 🐍 for Science 🔭
Story#
From Childhood to Code#
As a child, I began my journey into programming by developing software in VB6 and Delphi 7-Xe2. It was a thrilling hobby, allowing me to create GUI applications with ease and develop great softwares using these tools. However, these environments had their downsides. They often masked underlying complexities, and I didn’t learn much about algorithms or data structures. I was simply translating my ideas into code without truly understanding the mechanics. This approach wasn’t conducive to performance or a deep understanding of how things work. Learning C/C++ felt like a chore because it wasn’t as gratifying; development was slow, and the programs often looked ugly (console).
From Hobbies to Studies#
My academic journey led me to study Physics and Chemistry. Here, I delved into understanding how things work, from the basics of transistors to the principles of physics—what our ancient ancestors called « nature. » As I gained more confidence in my knowledge, I dabbled in Assembly 86x64 with AVX2, C, and C++. Despite my efforts to find the perfect programming language, success eluded me. Becoming a first-class developer requires an intimate understanding of architecture and more. Creating a Software as a Service (SaaS) with a Python backend and a JavaScript frontend doesn’t demand top-tier performance, but that isn’t my goal.
The Quest for Practicality#
My goal is to use my computer to solve physics problems and create small simulations and tools for my students. Python emerged as the ideal choice for this purpose. It’s versatile, easy to learn, and supported by a vast ecosystem of libraries suited for scientific computing. Python allows me to efficiently translate complex scientific concepts into code, providing a balance between ease of use and powerful capabilities.
Another small benchmark#
In this brief article, I want to illustrate the ease or difficulty of using common libraries and explain why I have decided to abandon C++ for my teaching activities as a physics teacher.
The bench#
For instance, consider the task of multiplying two 3000x3000 matrices, a common problem in scientific computing. In Python, using libraries like NumPy, this task becomes straightforward and efficient. See how this task is done in C++.
Beginner approach#
Python with Numpy#
import numpy as np
import time
# Create large matrices
a = np.random.rand(3000, 3000)
b = np.random.rand(3000, 3000)
# Time the matrix multiplication
start = time.time()
c = np.dot(a, b)
end = time.time()
print(f"Time taken by NumPy: {end - start} seconds")
Astuce
Results for Python / Numpy : 0.1469 seconds
C++ - Naïve - MSVC#
cmake_minimum_required(VERSION 3.28)
project(Simple)
set(CMAKE_CXX_STANDARD 17)
# Add optimization flags for Release build type
set(CMAKE_CXX_FLAGS_RELEASE "${CMAKE_CXX_FLAGS_RELEASE} /O2 /Ox")
add_executable(Simple main.cpp)
#include <iostream>
#include <vector>
#include <chrono>
void matmul(const std::vector<std::vector<double>>& A,
const std::vector<std::vector<double>>& B,
std::vector<std::vector<double>>& C) {
int n = A.size();
for (int i = 0; i < n; ++i) {
for (int j = 0; j < n; ++j) {
C[i][j] = 0;
for (int k = 0; k < n; ++k) {
C[i][j] += A[i][k] * B[k][j];
}
}
}
}
int main() {
int n = 3000;
std::vector<std::vector<double>> A(n, std::vector<double>(n));
std::vector<std::vector<double>> B(n, std::vector<double>(n));
std::vector<std::vector<double>> C(n, std::vector<double>(n));
// Initialize matrices
for (int i = 0; i < n; ++i) {
for (int j = 0; j < n; ++j) {
A[i][j] = rand() / double(RAND_MAX);
B[i][j] = rand() / double(RAND_MAX);
}
}
auto start = std::chrono::high_resolution_clock::now();
matmul(A, B, C);
auto end = std::chrono::high_resolution_clock::now();
std::chrono::duration<double> duration = end - start;
std::cout << "Time taken by C++: " << duration.count() << " seconds" << std::endl;
return 0;
}
Astuce
Results for C++ / Naïve : 55.9159 seconds
How to explain the differences between the two ?#
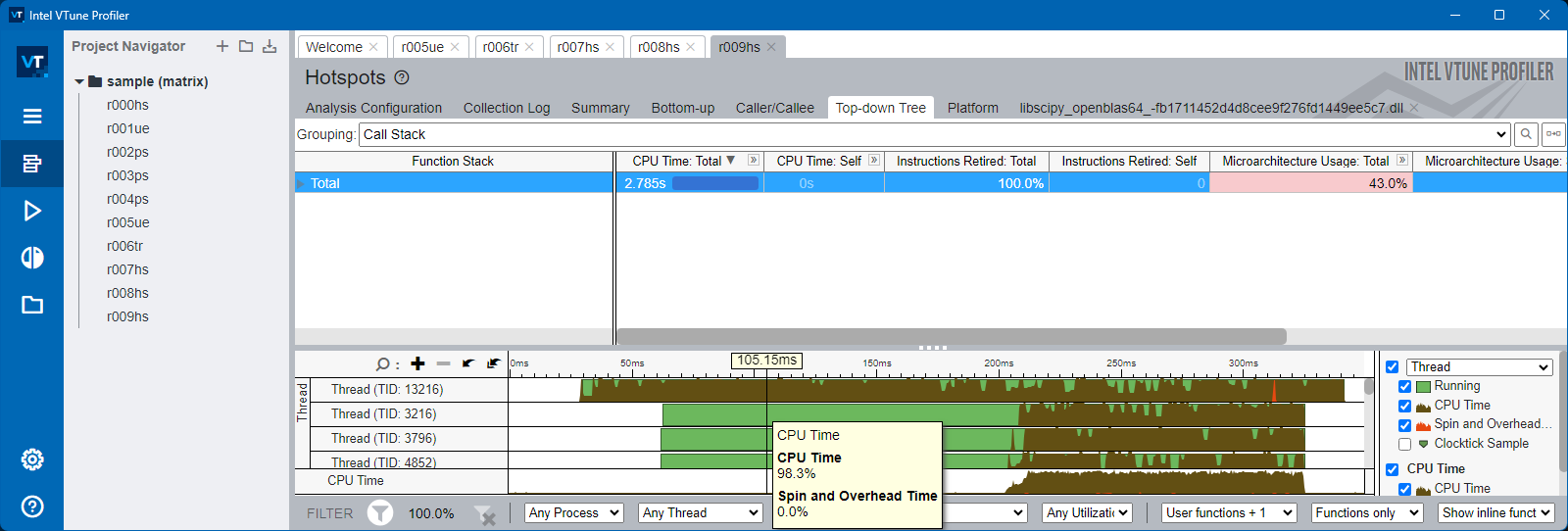
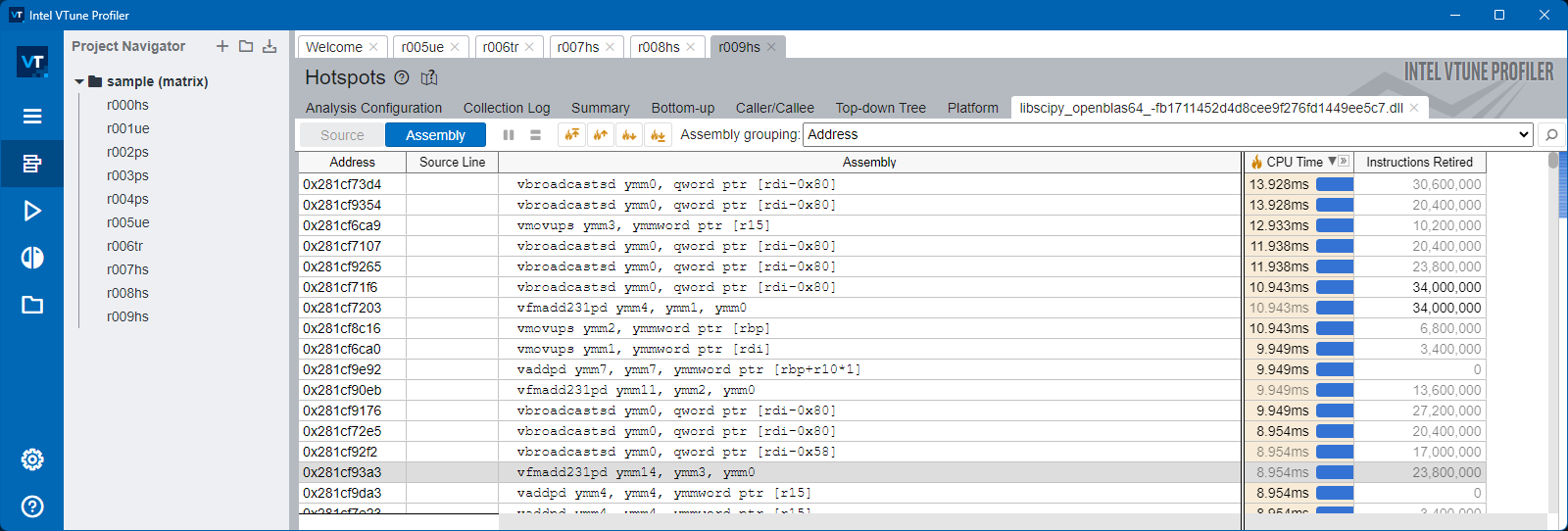
To inspect and study this, we will use Intel VTune profiler
Numpy#
C++ - MSVC#
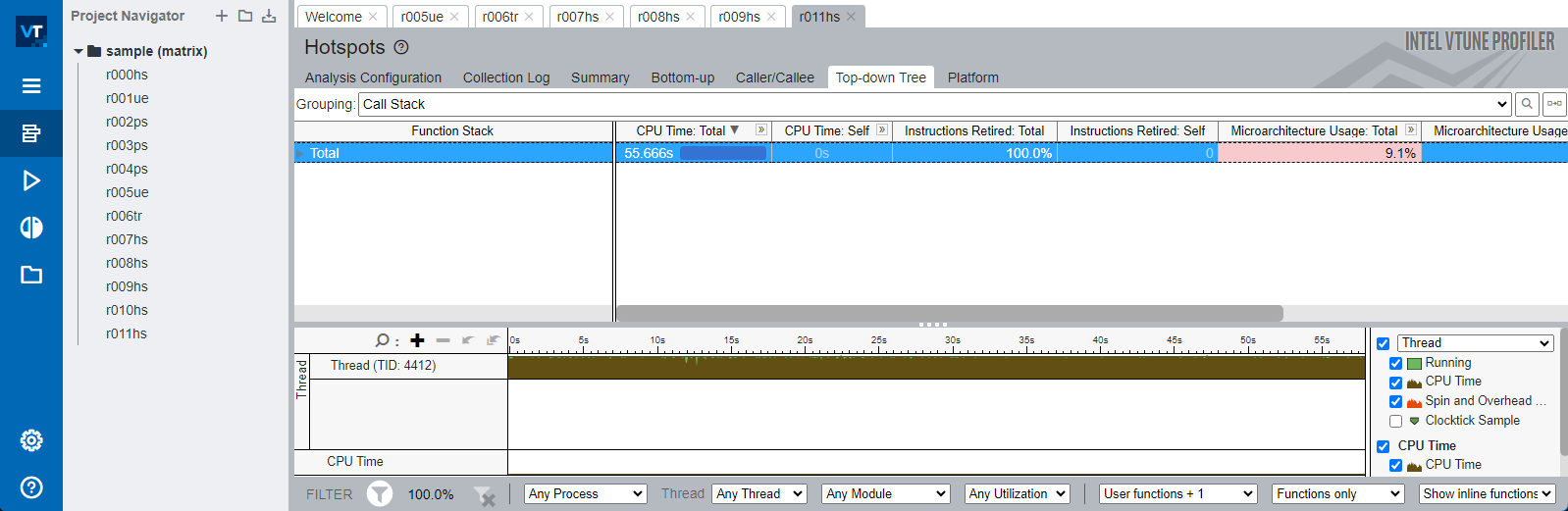
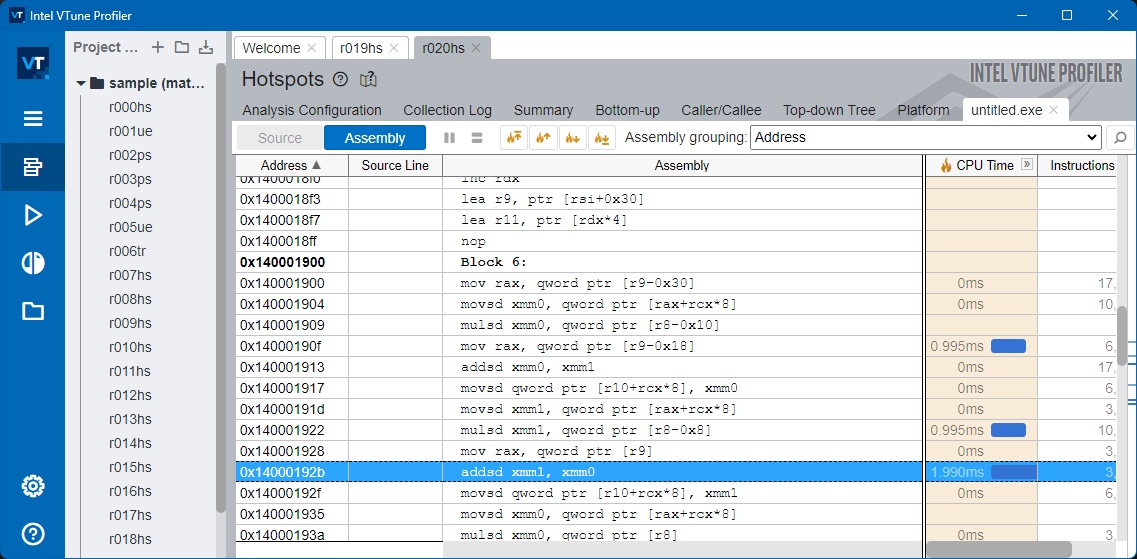
Danger
My Naïve version isn’t using AVX2 but SSE instruction set and isn’t multithreaded
Improving the C++ program#
AVX2 and Multithreading with OpenMP#
cmake_minimum_required(VERSION 3.28)
project(SimpleAVX2MP)
set(CMAKE_CXX_STANDARD 17)
# Add optimization flags for Release build type
if (MSVC)
set(CMAKE_CXX_FLAGS_RELEASE "${CMAKE_CXX_FLAGS_RELEASE} /O2 /Ox /Arch:AVX2")
endif()
# Find OpenMP
find_package(OpenMP REQUIRED)
add_executable(SimpleAVX2MP main.cpp)
# Link OpenMP libraries
if(OpenMP_CXX_FOUND)
target_link_libraries(SimpleAVX2MP PUBLIC OpenMP::OpenMP_CXX)
endif()
#include <iostream>
#include <vector>
#include <chrono>
#include <omp.h>
void matmul(const std::vector<std::vector<double>>& A,
const std::vector<std::vector<double>>& B,
std::vector<std::vector<double>>& C) {
int n = A.size();
#pragma omp parallel for
for (int i = 0; i < n; ++i) {
for (int j = 0; j < n; ++j) {
C[i][j] = 0;
for (int k = 0; k < n; ++k) {
C[i][j] += A[i][k] * B[k][j];
}
}
}
}
int main() {
int n = 3000;
std::vector<std::vector<double>> A(n, std::vector<double>(n));
std::vector<std::vector<double>> B(n, std::vector<double>(n));
std::vector<std::vector<double>> C(n, std::vector<double>(n));
// Initialize matrices
for (int i = 0; i < n; ++i) {
for (int j = 0; j < n; ++j) {
A[i][j] = rand() / double(RAND_MAX);
B[i][j] = rand() / double(RAND_MAX);
}
}
auto start = std::chrono::high_resolution_clock::now();
matmul(A, B, C);
auto end = std::chrono::high_resolution_clock::now();
std::chrono::duration<double> duration = end - start;
std::cout << "Time taken by C++: " << duration.count() << " seconds" << std::endl;
return 0;
}
Astuce
Results for C++ / OpenMP / AVX2 : 7.4244 seconds
Using a specialized library like Eigen#
cmake_minimum_required(VERSION 3.28)
project(EigenOpenMPAVX2)
set(CMAKE_CXX_STANDARD 17)
# Add optimization flags for Release build type
set(CMAKE_CXX_FLAGS_RELEASE "${CMAKE_CXX_FLAGS_RELEASE} /O2 /Ox /Ob3 /arch:AVX2")
find_package(OpenMP REQUIRED)
# Add the path to the Eigen directory
set(EIGEN3_INCLUDE_DIR "C:/Users/remi/CLionProjects/untitled1/eigen")
# Include the Eigen directory
include_directories(${EIGEN3_INCLUDE_DIR})
add_executable(EigenOpenMPAVX2 main.cpp)
if(OpenMP_CXX_FOUND)
target_link_libraries(EigenOpenMPAVX2 PUBLIC OpenMP::OpenMP_CXX)
add_definitions(-DEIGEN_USE_THREADS)
endif()
#include <iostream>
#include <Eigen/Dense>
#include <chrono>
#include <omp.h>
int main() {
int n = 3000;
Eigen::MatrixXd A = Eigen::MatrixXd::Random(n, n);
Eigen::MatrixXd B = Eigen::MatrixXd::Random(n, n);
Eigen::MatrixXd C(n, n);
//Set the number of threads for OpenMP
int num_threads = omp_get_max_threads();
omp_set_num_threads(num_threads);
Eigen::setNbThreads(num_threads);
std::cout << "Using " << num_threads << " threads for Eigen." << std::endl;
auto start = std::chrono::high_resolution_clock::now();
C.noalias() = A * B; // Use noalias to avoid temporary allocation
auto end = std::chrono::high_resolution_clock::now();
std::chrono::duration<double> duration = end - start;
std::cout << "Time taken by Eigen with OpenMP: " << duration.count() << " seconds" << std::endl;
return 0;
}
Astuce
Results for C++ / OpenMP / AVX2 / Eigen : 0.2219 seconds
It’s better but not as a fresh install of numpy (10 seconds to install it)
Using OpenBLAS with MinGW#
cmake_minimum_required(VERSION 3.15)
project(OpenBLASExample)
set(CMAKE_CXX_STANDARD 14)
# Define the path to your OpenBLAS installation
set(OpenBLAS_DIR "C:/Users/remi/CLionProjects/obl/openblas")
# Add the OpenBLAS include and library directories
include_directories(${OpenBLAS_DIR}/include)
link_directories(${OpenBLAS_DIR}/lib)
# Add an executable (replace main.cpp with your source file)
add_executable(OpenBLASExample main.cpp)
# Set compiler optimization flags
set(COMPILER_OPT_FLAGS "-O3 -march=native -flto -fopenmp")
# Link OpenBLAS library
target_link_libraries(OpenBLASExample ${OpenBLAS_DIR}/lib/libopenblas.a)
#include <cblas.h>
#include <iostream>
#include <chrono>
int main() {
const int size = 3000;
double *A = new double[size * size];
double *B = new double[size * size];
double *C = new double[size * size];
for (int i = 0; i < size * size; ++i) {
A[i] = rand() / double(RAND_MAX);
B[i] = rand() / double(RAND_MAX);
}
auto start = std::chrono::high_resolution_clock::now();
cblas_dgemm(CblasRowMajor, CblasNoTrans, CblasNoTrans, size, size, size, 1.0, A, size, B, size, 0.0, C, size);
auto end = std::chrono::high_resolution_clock::now();
std::chrono::duration<double> diff = end - start;
std::cout << "Time to multiply matrices with OpenBLAS: " << diff.count() << " s\n";
delete[] A;
delete[] B;
delete[] C;
return 0;
}
Astuce
Results for C++ / OpenMP / AVX2 / OpenBLAS : 0.1496 s
Using Intel Math Kernel Library#
cmake_minimum_required(VERSION 3.28)
project(MKLProject)
set(CMAKE_CXX_STANDARD 17)
# Add optimization flags for Release build type
set(CMAKE_CXX_FLAGS_RELEASE "${CMAKE_CXX_FLAGS_RELEASE} /O2 /Ox /Ob3 /arch:AVX2")
include_directories("C:/Program Files (x86)/Intel/oneAPI/compiler/2024.2/include")
link_directories("C:/Program Files (x86)/Intel/oneAPI/compiler/2024.2/lib")
# Include the MKL directories (adjust these paths according to your MKL installation)
include_directories("C:/Program Files (x86)/Intel/oneAPI/mkl/2024.2/include")
link_directories("C:/Program Files (x86)/Intel/oneAPI/mkl/2024.2/lib")
# Add executable
add_executable(MKLProject main.cpp)
# Link the MKL libraries
target_link_libraries(MKLProject PUBLIC mkl_intel_lp64 mkl_intel_thread mkl_core libiomp5md)
#include <iostream>
#include <chrono>
#include <mkl.h>
int main() {
int n = 3000;
// Allocate matrices
double* A = new double[n * n];
double* B = new double[n * n];
double* C = new double[n * n];
// Initialize matrices A and B with random values
for (int i = 0; i < n * n; ++i) {
A[i] = static_cast<double>(rand()) / RAND_MAX;
B[i] = static_cast<double>(rand()) / RAND_MAX;
C[i] = 0.0;
}
// Set the number of threads for MKL
int num_threads = mkl_get_max_threads();
mkl_set_num_threads(num_threads);
std::cout << "Using " << num_threads << " threads for MKL." << std::endl;
auto start = std::chrono::high_resolution_clock::now();
// Perform matrix multiplication using MKL's dgemm function
cblas_dgemm(CblasRowMajor, CblasNoTrans, CblasNoTrans, n, n, n, 1.0, A, n, B, n, 0.0, C, n);
auto end = std::chrono::high_resolution_clock::now();
std::chrono::duration<double> duration = end - start;
std::cout << "Time taken by MKL for matrix multiplication: " << duration.count() << " seconds" << std::endl;
// Clean up
delete[] A;
delete[] B;
delete[] C;
return 0;
}
Astuce
Results for C++ / Intel MKL : 0.1111 seconds
Working with Cython#
Naïve version#
from setuptools import setup
from Cython.Build import cythonize
setup(
ext_modules = cythonize("matrix_multiply.pyx")
)
import cython
from libc.stdlib cimport malloc, free, rand, RAND_MAX
from libc.time cimport clock, CLOCKS_PER_SEC
@cython.boundscheck(False)
@cython.wraparound(False)
cdef void matmul(double** A, double** B, double** C, int n):
cdef int i, j, k
for i in range(n):
for j in range(n):
C[i][j] = 0
for k in range(n):
C[i][j] += A[i][k] * B[k][j]
def matrix_multiply(int n):
cdef int i, j
cdef double** A = <double**>malloc(n * sizeof(double*))
cdef double** B = <double**>malloc(n * sizeof(double*))
cdef double** C = <double**>malloc(n * sizeof(double*))
cdef double start, end
for i in range(n):
A[i] = <double*>malloc(n * sizeof(double))
B[i] = <double*>malloc(n * sizeof(double))
C[i] = <double*>malloc(n * sizeof(double))
# Initialize matrices
for i in range(n):
for j in range(n):
A[i][j] = rand() / RAND_MAX
B[i][j] = rand() / RAND_MAX
start = clock()
matmul(A, B, C, n)
end = clock()
# Free allocated memory
for i in range(n):
free(A[i])
free(B[i])
free(C[i])
free(A)
free(B)
free(C)
return (end - start) / CLOCKS_PER_SEC
Astuce
Results for Cython : 60.487 seconds
With small optimization#
from setuptools import setup, Extension
from Cython.Distutils import build_ext
ext_modules = [
Extension(
'matrix_multiply2',
sources=['matrix_multiply2.pyx'],
extra_compile_args=['/openmp', '/O2','/arch:AVX2'] # Enable OpenMP and AVX2
)
]
setup(
name='matrix_multiply2',
ext_modules=ext_modules,
cmdclass={'build_ext': build_ext},
)
import cython
from libc.stdlib cimport malloc, free, rand, RAND_MAX
from libc.time cimport clock, CLOCKS_PER_SEC
from cython.parallel cimport prange, parallel
@cython.boundscheck(False)
@cython.wraparound(False)
cdef void matmul(double** A, double** B, double** C, int n):
cdef int i, j, k
# Use OpenMP to parallelize the outer loop
for i in prange(n, nogil=True,num_threads=16):
for j in range(n):
C[i][j] = 0
for k in range(n):
C[i][j] += A[i][k] * B[k][j]
def matrix_multiply(int n):
cdef int i, j
cdef double** A = <double**>malloc(n * sizeof(double*))
cdef double** B = <double**>malloc(n * sizeof(double*))
cdef double** C = <double**>malloc(n * sizeof(double*))
cdef double start, end
for i in range(n):
A[i] = <double*>malloc(n * sizeof(double))
B[i] = <double*>malloc(n * sizeof(double))
C[i] = <double*>malloc(n * sizeof(double))
# Initialize matrices
for i in range(n):
for j in range(n):
A[i][j] = rand() / RAND_MAX
B[i][j] = rand() / RAND_MAX
start = clock()
matmul(A, B, C, n)
end = clock()
# Free allocated memory
for i in range(n):
free(A[i])
free(B[i])
free(C[i])
free(A)
free(B)
free(C)
return (end - start) / CLOCKS_PER_SEC
Astuce
Results for Cython / OpenMP / AVX2 : 7.363 seconds
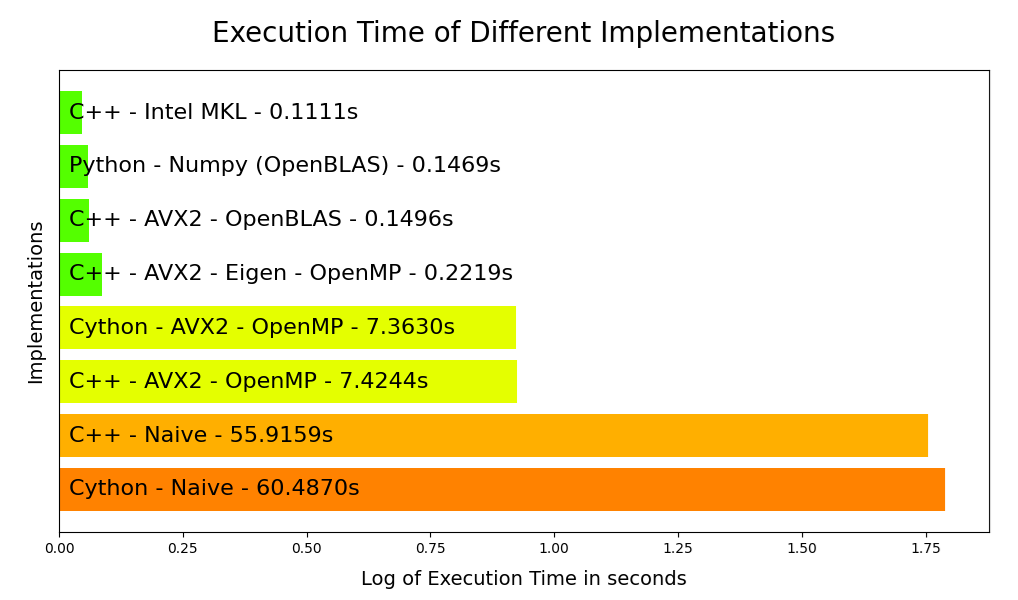
Benchmark results#
So to recap :
import matplotlib.pyplot as plt
import numpy as np
import matplotlib.colors as mcolors
# Data
languages = ['Cython - Naive', 'C++ - Naive', 'C++ - AVX2 - OpenMP', 'Cython - AVX2 - OpenMP',
'C++ - AVX2 - Eigen - OpenMP', 'C++ - AVX2 - OpenBLAS', 'Python - Numpy (OpenBLAS)', 'C++ - Intel MKL']
r_times = [60.487, 55.9159, 7.4244, 7.363, 0.2219, 0.1496, 0.1469, 0.1111]
times = [time + 1 for time in r_times]
times
log_times = np.log10(times)
plt.figure(figsize=(12, 6))
# Combine language names and times for the labels
labels = [f'{lang} - {time:.4f}s' for lang, time in zip(languages, r_times)]
# Create a colormap from red to blue
cmap = plt.get_cmap('prism_r')
norm = mcolors.Normalize(vmin=min(r_times), vmax=max(r_times))
colors = [cmap(norm(time)) for time in log_times]
# Create horizontal bar chart
bars = plt.barh(range(len(languages)), log_times, color=colors)
# Set the global font size to 14
plt.rcParams.update({'font.size': 14})
# Add text labels on the bars
for bar, label in zip(bars, labels):
plt.text(0.02, bar.get_y() + bar.get_height() / 2, label, va='center', ha='left', color='black', fontsize=16)
# Remove y-axis labels
plt.yticks([])
plt.xlabel('Log of Execution Time in seconds', fontsize=14, labelpad=10)
plt.ylabel('Implementations', fontsize=14, labelpad=10)
plt.title('Execution Time of Different Implementations', pad=20, fontsize=20)
plt.show()
Conclusion#
Clearly, the three lines of code for Numpy are unbeatable. Within a minute, I achieve performance levels that surpass anything I can produce in C++ (except when using Intel MKL, which is a proprietary library).
C++ allows for complete, meticulous control over what you execute on the processor. However, using a library like OpenBLAS or Intel MKL is akin to using a black box, just like Numpy.
Talented scientists and engineers have worked on these libraries. Why reinvent the wheel? Why start from scratch as a scientist?
Python is the perfect tool because it is simple and intuitive for teaching physics. Its performance, when used correctly (with compiled libraries for critical parts), is more than sufficient for any university professor, at least up to a Master’s level.
